Get Started in 10 Minutes¶
Altay Sansal
Oct 20, 2025
9 min read
In this page we will be showing basic capabilities of MDIO.
For demonstration purposes, we will ingest the remote Teapot Dome open-source dataset. The dataset details and licensing can be found at the SEG Wiki.
We are using the 3D seismic stack dataset named filt_mig.sgy.
The full HTTP link for the dataset (hosted on AWS): http://s3.amazonaws.com/teapot/filt_mig.sgy
Warning
For plotting and remote ingestion the notebook requires Matplotlib and aiohttp
as a dependency. Please install it before executing using pip install matplotlib aiohttp or
conda install matplotlib aiohttp.
Defining the SEG-Y Dataset¶
Since MDIO 0.8 we can directly ingest remote SEG-Y files! The file is 386 MB in size. To make the header scan performant we can also set up an environment variable for MDIO. See here for when to use this: Buffered Reads in Ingestion.
The dataset is irregularly shaped, however it is padded to a rectangle with zeros (dead traces). We will see that later at the live mask plotting.
The following environment variables are essential here:
MDIO__IMPORT__CLOUD_NATIVEtells MDIO to do buffered reads for headers due to remote file.MDIO__IMPORT__SAVE_SEGY_FILE_HEADERmakes MDIO save the SEG-Y specific file headers (text, binary) which is not strictly necessary for consumption and is disabled by default.
import os
os.environ["MDIO__IMPORT__CLOUD_NATIVE"] = "true"
os.environ["MDIO__IMPORT__SAVE_SEGY_FILE_HEADER"] = "true"
input_url = "http://s3.amazonaws.com/teapot/filt_mig.sgy"
Ingesting to MDIO¶
To do this, we can use the convenient SEG-Y to MDIO converter.
The inline and crossline values are located at bytes 181 and 185. Note that this doesn’t match any SEG-Y standards.
MDIO uses TGSAI/segy to parse the SEG-Y; the field names conform to its canonical keys defined in SEGY Binary Header Keys and SEGY Trace Header Keys. Since MDIO v1 we also introduced templates for common seismic data types. For instance, we will be using the PostStack3DTime template here, which expects the same canonical keys.
We will also specify the units for the time domain. The spatial units will be automatically parsed from SEG-Y binary header. However, there may be a case where it is corrupt in the file, for that see the Fixing X/Y Units Issues section.
In summary, we will use the byte locations as defined for ingestion.
import matplotlib.pyplot as plt
from segy.schema import HeaderField
from segy.standards import get_segy_standard
from mdio import segy_to_mdio
from mdio.builder.schemas.v1.units import TimeUnitModel
from mdio.builder.template_registry import get_template
teapot_trace_headers = [
HeaderField(name="inline", byte=181, format="int32"),
HeaderField(name="crossline", byte=185, format="int32"),
HeaderField(name="cdp_x", byte=189, format="int32"),
HeaderField(name="cdp_y", byte=193, format="int32"),
]
rev0_segy_spec = get_segy_standard(0)
teapot_segy_spec = rev0_segy_spec.customize(trace_header_fields=teapot_trace_headers)
mdio_template = get_template("PostStack3DTime")
unit_ms = TimeUnitModel(time="ms")
mdio_template.add_units({"time": unit_ms})
segy_to_mdio(
input_path=input_url,
output_path="filt_mig.mdio",
segy_spec=teapot_segy_spec,
mdio_template=mdio_template,
overwrite=True,
)
It only took a few seconds to ingest, since this is a very small file.
However, MDIO scales up to TB (that’s ~1,000 GB) sized volumes!
Opening the Ingested MDIO File¶
Let’s open the MDIO file with the open_mdio function. This will return a pretty xarray representation with our standardized format.
from mdio import open_mdio
dataset = open_mdio("filt_mig.mdio")
dataset
<xarray.Dataset> Size: 403MB
Dimensions: (inline: 345, crossline: 188, time: 1501)
Coordinates:
* inline (inline) int32 1kB 1 2 3 4 5 6 ... 340 341 342 343 344 345
* crossline (crossline) int32 752B 1 2 3 4 5 6 ... 184 185 186 187 188
* time (time) int32 6kB 0 2 4 6 8 10 ... 2992 2994 2996 2998 3000
cdp_y (inline, crossline) float64 519kB ...
cdp_x (inline, crossline) float64 519kB ...
Data variables:
trace_mask (inline, crossline) bool 65kB ...
segy_file_header <U1 4B ...
amplitude (inline, crossline, time) float32 389MB ...
headers (inline, crossline) [('trace_seq_num_line', '<i4'), ('trace_seq_num_reel', '<i4'), ('orig_field_record_num', '<i4'), ('trace_num_orig_record', '<i4'), ('energy_source_point_num', '<i4'), ('ensemble_num', '<i4'), ('trace_num_ensemble', '<i4'), ('trace_id_code', '<i2'), ('vertically_summed_traces', '<i2'), ('horizontally_stacked_traces', '<i2'), ('data_use', '<i2'), ('source_to_receiver_distance', '<i4'), ('receiver_group_elevation', '<i4'), ('source_surface_elevation', '<i4'), ('source_depth_below_surface', '<i4'), ('receiver_datum_elevation', '<i4'), ('source_datum_elevation', '<i4'), ('source_water_depth', '<i4'), ('receiver_water_depth', '<i4'), ('elevation_depth_scalar', '<i2'), ('coordinate_scalar', '<i2'), ('source_coord_x', '<i4'), ('source_coord_y', '<i4'), ('group_coord_x', '<i4'), ('group_coord_y', '<i4'), ('coordinate_unit', '<i2'), ('weathering_velocity', '<i2'), ('subweathering_velocity', '<i2'), ('source_uphole_time', '<i2'), ('group_uphole_time', '<i2'), ('source_static_correction', '<i2'), ('receiver_static_correction', '<i2'), ('total_static_applied', '<i2'), ('lag_time_a', '<i2'), ('lag_time_b', '<i2'), ('delay_recording_time', '<i2'), ('mute_time_start', '<i2'), ('mute_time_end', '<i2'), ('samples_per_trace', '<i2'), ('sample_interval', '<i2'), ('instrument_gain_type', '<i2'), ('instrument_gain_const', '<i2'), ('instrument_gain_initial', '<i2'), ('correlated_data', '<i2'), ('sweep_freq_start', '<i2'), ('sweep_freq_end', '<i2'), ('sweep_length', '<i2'), ('sweep_type', '<i2'), ('sweep_taper_start', '<i2'), ('sweep_taper_end', '<i2'), ('taper_type', '<i2'), ('alias_filter_freq', '<i2'), ('alias_filter_slope', '<i2'), ('notch_filter_freq', '<i2'), ('notch_filter_slope', '<i2'), ('low_cut_freq', '<i2'), ('high_cut_freq', '<i2'), ('low_cut_slope', '<i2'), ('high_cut_slope', '<i2'), ('year_recorded', '<i2'), ('day_of_year', '<i2'), ('hour_of_day', '<i2'), ('minute_of_hour', '<i2'), ('second_of_minute', '<i2'), ('time_basis_code', '<i2'), ('trace_weighting_factor', '<i2'), ('group_num_roll_switch', '<i2'), ('group_num_first_trace', '<i2'), ('group_num_last_trace', '<i2'), ('gap_size', '<i2'), ('taper_overtravel', '<i2'), ('inline', '<i4'), ('crossline', '<i4'), ('cdp_x', '<i4'), ('cdp_y', '<i4')] 13MB ...
Attributes:
apiVersion: 1.0.8
createdOn: 2025-10-20 15:52:03.112832+00:00
name: PostStack3DTime
attributes: {'surveyType': '3D', 'gatherType': 'stacked', 'defaultVariab...Querying Metadata¶
Now let’s look at the file text header saved in the segy_file_header metadata variable.
print(dataset["segy_file_header"].attrs["textHeader"])
C 1 CLIENT: ROCKY MOUNTAIN OILFIELD TESTING CENTER
C 2 PROJECT: NAVAL PETROLEUM RESERVE #3 (TEAPOT DOME); NATRONA COUNTY, WYOMING
C 3 LINE: 3D
C 4
C 5 THIS IS THE FILTERED POST STACK MIGRATION
C 6
C 7 INLINE 1, XLINE 1: X COORDINATE: 788937 Y COORDINATE: 938845
C 8 INLINE 1, XLINE 188: X COORDINATE: 809501 Y COORDINATE: 939333
C 9 INLINE 188, XLINE 1: X COORDINATE: 788039 Y COORDINATE: 976674
C10 INLINE NUMBER: MIN: 1 MAX: 345 TOTAL: 345
C11 CROSSLINE NUMBER: MIN: 1 MAX: 188 TOTAL: 188
C12 TOTAL NUMBER OF CDPS: 64860 BIN DIMENSION: 110' X 110'
C13
C14
C15
C16
C17
C18
C19 GENERAL SEGY INFORMATION
C20 RECORD LENGHT (MS): 3000
C21 SAMPLE RATE (MS): 2.0
C22 DATA FORMAT: 4 BYTE IBM FLOATING POINT
C23 BYTES 13- 16: CROSSLINE NUMBER (TRACE)
C24 BYTES 17- 20: INLINE NUMBER (LINE)
C25 BYTES 81- 84: CDP_X COORD
C26 BYTES 85- 88: CDP_Y COORD
C27 BYTES 181-184: INLINE NUMBER (LINE)
C28 BYTES 185-188: CROSSLINE NUMBER (TRACE)
C29 BYTES 189-192: CDP_X COORD
C30 BYTES 193-196: CDP_Y COORD
C31
C32
C33
C34
C35
C36 Processed by: Excel Geophysical Services, Inc.
C37 8301 East Prentice Ave. Ste. 402
C38 Englewood, Colorado 80111
C39 (voice) 303.694.9629 (fax) 303.771.1646
C40 END EBCDIC
Since we saved the binary header, we can look at that as well.
dataset["segy_file_header"].attrs["binaryHeader"]
{'job_id': 9999,
'line_num': 9999,
'reel_num': 1,
'data_traces_per_ensemble': 188,
'aux_traces_per_ensemble': 0,
'sample_interval': 2000,
'orig_sample_interval': 0,
'samples_per_trace': 1501,
'orig_samples_per_trace': 1501,
'data_sample_format': 1,
'ensemble_fold': 57,
'trace_sorting_code': 4,
'vertical_sum_code': 1,
'sweep_freq_start': 0,
'sweep_freq_end': 0,
'sweep_length': 0,
'sweep_type_code': 0,
'sweep_trace_num': 0,
'sweep_taper_start': 0,
'sweep_taper_end': 0,
'taper_type_code': 0,
'correlated_data_code': 2,
'binary_gain_code': 1,
'amp_recovery_code': 4,
'measurement_system_code': 2,
'impulse_polarity_code': 1,
'vibratory_polarity_code': 0,
'segy_revision_major': 0,
'segy_revision_minor': 0}
Fetching Data and Plotting¶
Now we will demonstrate getting an inline from MDIO.
Since MDIO v1 we are using Xarray under the hood, so we can use its convenient indexing. It also handles the plotting with proper dimension coordinate labels.
MDIO stores summary statistics. We can calculate the standard deviation (std) value of the dataset to adjust the gain.
from mdio.builder.schemas.v1.stats import SummaryStatistics
stats = SummaryStatistics.model_validate_json(dataset["amplitude"].attrs["statsV1"])
std = ((stats.sum_squares / stats.count) - (stats.sum / stats.count) ** 2) ** 0.5
il_dataset = dataset.sel(inline=278)
il_amp = il_dataset["amplitude"].T
il_amp.plot(vmin=-2 * std, vmax=2 * std, cmap="gray_r", yincrease=False);
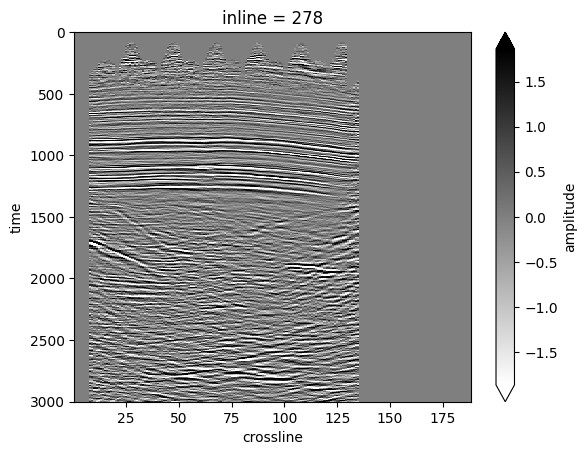
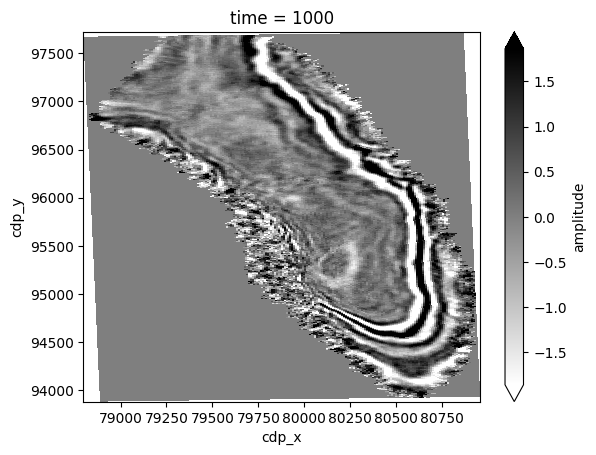
Let’s do the same with a time slice.
We will display two-way-time at 1,000 ms.
Note that since we parse the X/Y coordinates, we can plot time slice in real world coordinates.
twt_data = dataset["amplitude"].sel(time=1000)
twt_data.plot(vmin=-2 * std, vmax=2 * std, cmap="gray_r", x="cdp_x", y="cdp_y");
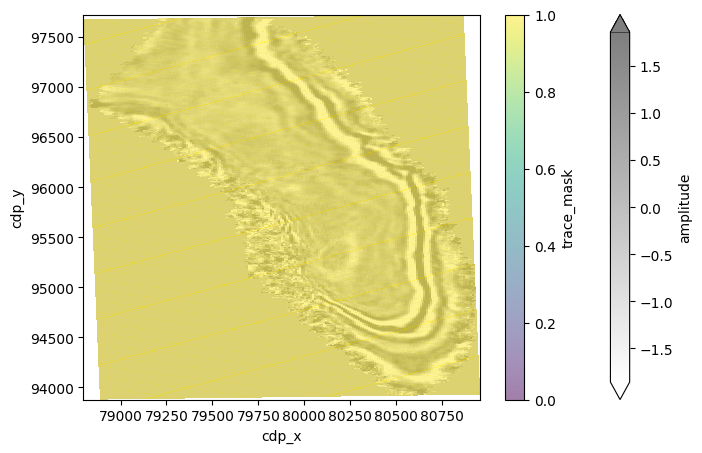
We can also overlay live mask with the time slice. However, in this example, the dataset is zero-padded.
The live trace mask will always show True (yellow).
trace_mask = dataset.trace_mask[:]
twt_data.plot(vmin=-2 * std, vmax=2 * std, cmap="gray_r", x="cdp_x", y="cdp_y", alpha=0.5, figsize=(8, 5))
trace_mask.plot(vmin=0, vmax=1, x="cdp_x", y="cdp_y", alpha=0.5);
Query Headers¶
We can query headers for the whole dataset very quickly because they are separated from the seismic wavefield.
Let’s get all the headers for X and Y coordinates in this dataset.
Note that the header maps will still share the geometry/grid of the dataset!
The compute property fetches the lazily opened MDIO values.
dataset.headers["cdp_x"].compute()
<xarray.DataArray 'cdp_x' (inline: 345, crossline: 188)> Size: 519kB
array([[78893.7, 78904.7, 78915.7, ..., 80928.2, 80939.2, 80950.2],
[78893.5, 78904.5, 78915.5, ..., 80927.9, 80938.9, 80949.9],
[78893.2, 78904.2, 78915.2, ..., 80927.6, 80938.6, 80949.6],
...,
[78804.4, 78815.4, 78826.4, ..., 80838.9, 80849.9, 80860.9],
[78804.2, 78815.2, 78826.2, ..., 80838.6, 80849.6, 80860.6],
[78803.9, 78814.9, 78825.9, ..., 80838.3, 80849.3, 80860.3]],
shape=(345, 188))
Coordinates:
* inline (inline) int32 1kB 1 2 3 4 5 6 7 ... 339 340 341 342 343 344 345
* crossline (crossline) int32 752B 1 2 3 4 5 6 7 ... 183 184 185 186 187 188
cdp_y (inline, crossline) float64 519kB 9.388e+04 ... 9.772e+04
cdp_x (inline, crossline) float64 519kB 7.889e+04 ... 8.086e+04
Attributes:
unitsV1: {'length': 'ft'}dataset.headers["cdp_y"].compute()
<xarray.DataArray 'cdp_y' (inline: 345, crossline: 188)> Size: 519kB
array([[93884.6, 93884.8, 93885.1, ..., 93932.9, 93933.1, 93933.4],
[93895.6, 93895.8, 93896.1, ..., 93943.9, 93944.1, 93944.4],
[93906.6, 93906.8, 93907.1, ..., 93954.9, 93955.1, 93955.4],
...,
[97645.5, 97645.8, 97646. , ..., 97693.8, 97694.1, 97694.3],
[97656.5, 97656.8, 97657. , ..., 97704.8, 97705.1, 97705.3],
[97667.5, 97667.8, 97668. , ..., 97715.8, 97716.1, 97716.3]],
shape=(345, 188))
Coordinates:
* inline (inline) int32 1kB 1 2 3 4 5 6 7 ... 339 340 341 342 343 344 345
* crossline (crossline) int32 752B 1 2 3 4 5 6 7 ... 183 184 185 186 187 188
cdp_y (inline, crossline) float64 519kB 9.388e+04 ... 9.772e+04
cdp_x (inline, crossline) float64 519kB 7.889e+04 ... 8.086e+04
Attributes:
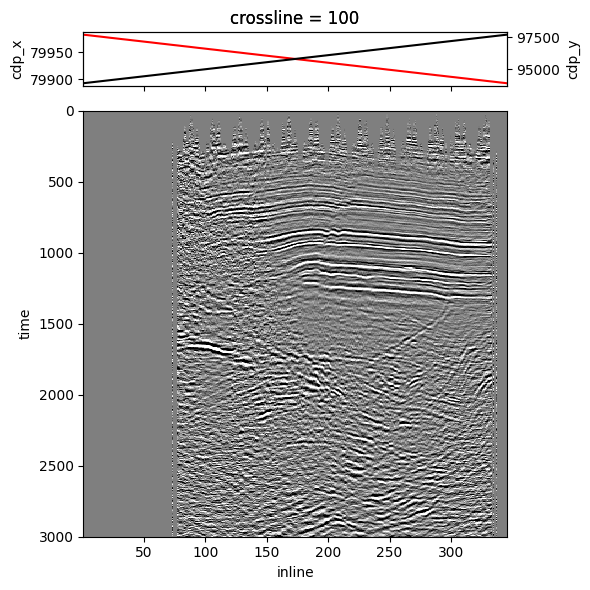
unitsV1: {'length': 'ft'}As we mentioned before, we can also get specific dataset slices of headers while fetching a slice.
Let’s fetch a crossline; we are still using some previous parameters.
Since the sliced dataset contains the headers as well, we can plot the headers on top of the image.
Full headers can be mapped and plotted as well, but we won’t demonstrate that here.
xl_dataset = dataset.sel(crossline=100) # slices everything available in MDIO dataset!
cdp_x_header = xl_dataset["cdp_x"]
cdp_y_header = xl_dataset["cdp_y"]
image = xl_dataset["amplitude"].T
# Build plot from here
gs_kw = {"height_ratios": (1, 8)}
fig, (hdr_ax, img_ax) = plt.subplots(2, 1, figsize=(6, 6), gridspec_kw=gs_kw, sharex="all")
hdr_ax2 = hdr_ax.twinx()
cdp_x_header.plot(ax=hdr_ax, c="red")
cdp_y_header.plot(ax=hdr_ax2, c="black")
image.plot(ax=img_ax, vmin=-2 * std, vmax=2 * std, cmap="gray_r", yincrease=False, add_colorbar=False)
img_ax.set_title("")
hdr_ax.set_xlabel("")
plt.tight_layout()
MDIO to SEG-Y Conversion¶
Finally, let’s demonstrate going back to SEG-Y.
We will use the convenient mdio_to_segy function and write it out as a round-trip file.
The output spec can be modified if we want to write things to different byte locations, etc, but we will use the same one as before.
from mdio import mdio_to_segy
mdio_to_segy(
input_path="filt_mig.mdio",
output_path="filt_mig_roundtrip.sgy",
segy_spec=teapot_segy_spec,
)
Validate Round-Trip SEG-Y File¶
We can validate if the round-trip SEG-Y file is matching the original using TGSAI/segy.
Step by step:
Open original file
Open round-trip file
Compare text headers
Compare binary headers
Compare 100 random headers and traces
import numpy as np
from segy import SegyFile
original_segy = SegyFile(input_url)
roundtrip_segy = SegyFile("filt_mig_roundtrip.sgy")
# Compare text header
assert original_segy.text_header == roundtrip_segy.text_header
# Compare bin header
assert original_segy.binary_header == roundtrip_segy.binary_header
# Compare 100 random trace headers and traces
rng = np.random.default_rng()
rand_indices = rng.integers(low=0, high=original_segy.num_traces, size=100)
for idx in rand_indices:
np.testing.assert_equal(original_segy.trace[idx], roundtrip_segy.trace[idx])
print("Files identical!")
Files identical!